| |
The same genetic blueprint gives rise to thousands of cell types that make up the human body. Intricate mechanisms govern the choice to make skin, heart, or brain cells. These different cell types must be correctly arranged in spatial patterns to make functioning tissues and organs. In many organisms with continual turnover of cells, the genome faces the additional challenge of ensuring the faithful transmission of information throughout a lifetime-over decades in the case of humans. Thus, how one genome encodes thousands of patterns in space and time is of central importance to biology and medicine. Inappropriate activation of genes can give rise to birth defects, premature aging, or cancer, among many other diseases. Restoration of proper organ function often requires restoring homeostatic gene regulation.
Long Noncoding RNAs and Positional Identity
As a practicing dermatologist, I am fascinated by what makes human skin from different parts of the body different, a fact that guides the diagnosis and treatment of many skin diseases. Why do long hairs grow on the scalp but not on our palms or soles? How do cells know where they are located in the body, and how do they remember this information?
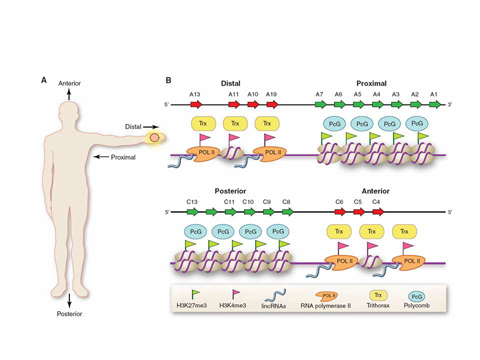
We discovered that one class of skin cells, the fibroblasts, encode the positional identity of skin via specific markings on their chromatin, the DNA-protein complex where genes reside. Based on the chromatin configurations of specific genes, most notably the HOX genes, fibroblasts differentially activate hundreds of genes based on the cell's location along three anatomic axes-anterior-posterior (head to tail), proximal-distal (close or far away from the trunk), and dermal-nondermal (surface or internal organ). This in effect creates a global positioning system for all cells to navigate.
These studies also revealed a surprising abundance of long intergenic noncoding RNAs (also known as lincRNAs, a newly recognized type of genes that do not encode proteins) that are involved in programming chromatin states. We are particularly fascinated by HOTAIR, the first known lincRNA that can regulate the chromatin state of genes on distantly located chromosomes. We now appreciate that the genome is pervasively transcribed to give rise to thousands of lincRNAs, which are likely to play key roles in the gene regulation of diverse biological states and disease. We are interested in understanding how lincRNAs control gene activity, and in deciphering the rules that will allow the functions of thousands of lincRNAs to be predicted and studied.
Chromatin as GPS device
Large-Scale Gene Regulatory Programs in Cancer Metastasis and Self-Renewal
In contrast to the orderly acquisition of positional identity, cancer progression is characterized by abrogation of normal positional boundaries, especially in metastasis, which is the leading cause of cancer death. We and many others have previously identified gene expression signatures (GES), composed of dozens to hundreds of genes, that distinguish indolent human cancers from those prone to metastasis; these signatures can provide improved prognostic prediction for cancer patients. Furthermore, we have developed methods to pinpoint master regulators of GES-singular control points that can toggle the activity of the entire genetic program. This allows complex gene programs observed in human cancers to be easily recapitulated in the laboratory as models for drug development. This has enabled the creation of faithful laboratory models of human cancer types, identified specific drugs that can target these cancers, and revealed the hierarchy of transcriptional programs involved in the generation of cancer stem cellscellsthe cells that continually repopulate a tumor or its metastases.
Aging: Genome Regulation over Time
Aging and the control of longevity have proved a difficult problem from the perspective of gene regulation. This is because different genes seemed to be turned on with age in different organs and animals, which has led to the view that aging is a result of stochastic breakdown of regulatory programs from passive wear and tear. We showed, however, that although the genes that change with age may be different throughout the body or in evolution, the master switches that turn on these different genes are actually conserved. The transcription factor NF-kB is a dominant regulator of age-associated GES in mammalian tissues, and inducible blockade of NF-kB in already aged mice can revert the tissue characteristics and its gene expression program to its youthful state. Nutrient availability is an important environmental influence on the rate of aging, and we recently found that the NF-kB pathway is the key output regulated by nutrient-sensing longevity regulators called sirtuins. These findings support the idea that aging is not just passive wear and tear, but is also enforced by an active genetic program that may be reversed for healthful benefits. Our ongoing efforts are focused on understanding how NF-kB activity becomes activated over organismal life, and why the consequences of NF-kB activation change with time.
|

